Protein-protein Interactions
New Technologies to detect, modulate, and functionally analyze Protein-Protein Interactions
The human protein “interactome” may involve ~130,000 to ~650,000 protein-protein interactions (PPIs). The mutations that lead to cancer often perturb PPIs and as a consequence change signal transduction pathways that regulate cell growth, death, control of genetic stability, interaction between cells, etc . Currently, a critical unmet need is the availability of real time assays that directly measure PPIs in living cells. Such an assay would enable the consequences of cancer associated mutations to perturb specific PPIs, and to evaluate their consequences on diverse biologic processes relevant to cancer and other diseases.
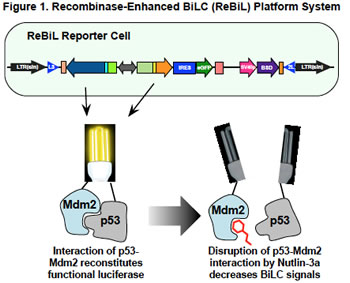 Equally importantly, the availability of robust, rapid, and specific methods to detect PPIs would enable the development of high throughput screens to identify and validate drugs with the ability to disrupt protein complexes in living cells. To address this need, we developed a plug-and-play platform system to detect PPIs in cells and in cell lysates. This system integrates Cre-recombinase mediated cassette exchange (RMCE) and bi-molecular luciferase complementation (BiLC). We have named this platform system ReBiL, Recombinase-enhanced BiLC (ReBiL) (Figure 1).
Equally importantly, the availability of robust, rapid, and specific methods to detect PPIs would enable the development of high throughput screens to identify and validate drugs with the ability to disrupt protein complexes in living cells. To address this need, we developed a plug-and-play platform system to detect PPIs in cells and in cell lysates. This system integrates Cre-recombinase mediated cassette exchange (RMCE) and bi-molecular luciferase complementation (BiLC). We have named this platform system ReBiL, Recombinase-enhanced BiLC (ReBiL) (Figure 1).
ReBiL enables detection of PPIs that cannot be detected by standard methods, such as transient transfection. As one example, we compared the ability of ReBiL and a transient transfection to detect interaction between proteins that have low affinity for each other, and interact very transiently. The test system involved interaction between the E2 conjugating protein Ube2t and the E3 ubiquitin ligase FANCL. These proteins are critical parts of the Fanconi Anemia complex that participates in repair of damaged DNA. Mutations in this pathway cause hematological abnormalities and create a predisposition to develop cancer. We showed that transient transfection of Ube2t and FANCL split luciferase fusions did not detect their interaction, while ReBiL employing the same constructs produced a robust and readily reproducible signal. We could not detect a signal from a PPI pair encoding a mutation that prevents Ube2t-FANCL interaction. This demonstrates the high sensitivity and specificity of the ReBiL platform.
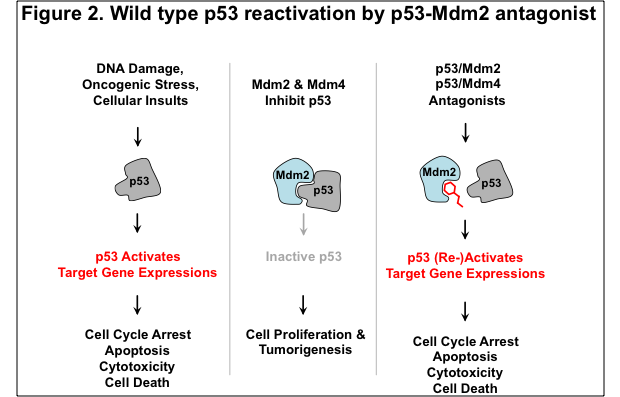
We next turned our attention to determining whether the ReBiL platform could be used to analyze drug candidates useful to treat cancer. Here, we analyzed a number of drugs that are being developed to treat cancers that have a wild type p53 protein, but a defective p53 tumor suppressor pathway due to overproduction of one of its negative regulators, Mdm2 or Mdm4 (see Figure2). The p53 pathway controls the response of cells to myriad conditions that impinge on cell viability and faithful replication and segregation of DNA, including DNA damage, nutrient availability, various metabolic stresses, activation of oncogenes, etc. It is also one of the most frequently mutated pathways in cancer, since its inactivation enables cells to proliferate under conditions that can generate and perpetuate unstable genomes.
As ReBiL enables the rapid generation and insertion of BiLC fusion constructs into the genome of specially engineered reporter cells, we developed cell lines optimized to detect the activities of drugs designed to interfere with p53-Mdm2, p53-Mdm4 and Mdm2-Mdm4 interactions. We found that reported small molecule inhibitors of the interaction between p53 and Mdm2 work as predicted in both cell lysates and in living cells. By contrast, we were able to show for the first time that stapled peptides work in cell lysates, but they do not get into cells efficiently. We were surprised to find that the p53 agonist stapled peptides that were optimized for cell entry actually ended up killing normal and cancer cells. We also used ReBiL to rapidly assess the mechanism of cell killing, and we found that this is a p53-independent process that is antagonized by serum. We have also shown that ReBiL can be used for high throughput robotic screening of a two million compound library in 1536-well plate format.
The ability to use ReBiL to rapidly clone and analyze interaction between different mutants from the same chromosomal locus, and to determine the amount of protein expressed through doxycycline mediated induction has proven to be of enormous importance. For example, we have now extended the use of ReBiL to investigate the mechanisms by which wild type and oncogenic Ras proteins signal. These studies are revealing the importance of membrane association for Ras signaling, and they are enabling us to refine the domains of Ras that are essential. Furthermore, they enable us to interrogate the potential of other Ras-interacting molecules to modulate Ras signaling via protein-protein and protein-membrane interactions.
Taken together, these observations establish ReBiL as a broad based platform system useful for identifying and analyzing diverse protein interactions that vary in their strength and duration of association and subcellular localization. ReBiL provides a convenient and cost-effective strategy for high-throughput screening of large compound libraries, which has implications for cancer and other diseases of great medical significance.
Useful WebLinks
The ReBiL platform technology is available for licensinghttp://salkinnovations.testtechnologypublisher.com/technology/7022
The p53 mutation database
http://www-p53.iarc.fr/index.html
p53, Mdm2 and Mdmx-related information
http://p53.free.fr/index.html
American Association for Cancer Research
http://www.aacr.org
Personnel
Yao-Cheng (Leo) Li, Ph.D. graduated with a B. S. in Plant Pathology, 1992 from National Chung-Hsing University and a M.S. in Biochemistry, 1994 from the National Yang-Ming University, Taiwan. He graduated in 2002 with a Ph.D. in Molecular and Cellular Pharmacology from the Rutgers, the State University of New Jersey and Robert Wood Johnson Medical School, New Jersey. Leo joined the Wahl Lab in 2003.